What the Farkle? Recursion, Modeling, and Genetic Algorithms with KNIME, Part 4
Welcome to the fourth and final entry in my series on the dice game called Farkle. In parts one and two I covered the basic rules of the game and how to model player strategy using KNIME. The basic model that I developed in those two entries led me to use a genetic algorithm to evolve player strategy. As a brief review, a genetic algorithm is a search algorithm to efficiently find an answer to a problem that does not lend itself to an exhaustive analysis. A genetic algorithm uses the notions of Charles Darwin’s theory of evolution in five phases:
- Initial population
- Fitness function
- Selection
- Crossover
- Mutation
In part three, I covered the first three steps of the algorithm that I built using a workflow that is available on the KNIME hub. The workflow starts with an initial set of 400 random player strategies and the scores them with a Monte Carlo simulation of a large number of individual turns for each player. The players with the top ten average scores are then selected for the final two steps of the algorithm: crossover and mutation. The result of the final two steps is a new generation of players that can then be run through the same algorithm, ideally resulting in an overall improvement of the average score of the new top ten players. This process is repeated until there is no appreciable increase in the value of the fitness function between generations.
In my implementation of a genetic algorithm, crossover is done by randomly combining each of the top ten players with each other. Furthermore, four offspring are created from each player pair so that different combinations of each pair of parents’ attributes can be considered. This also has the nice side effect of producing a new generation that is the same size as the previous generation. The potential downside of this somewhat arbitrary selection and crossover configuration is that the scope of player selection and consideration of various players combinations is not broad enough and results in positive behaviors that do not survive in the population.
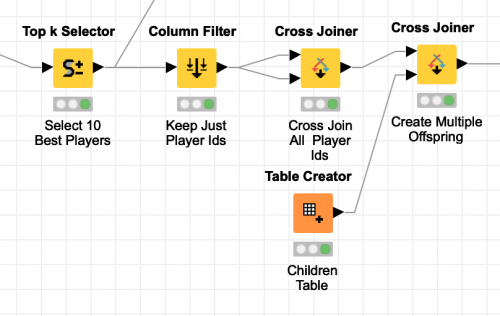
Producing every combination of the top 10 players is easy in KNIME thanks to the Cross Joiner node. Just like a database cross join, the output of this node is every combination of the rows from its two input tables. By supplying the same table of player ids as both inputs to the node, all possible permutations of the top 10 players are output. The node even allows me to configure a suffix for the column names of the second table so that we end up with two uniquely named columns:
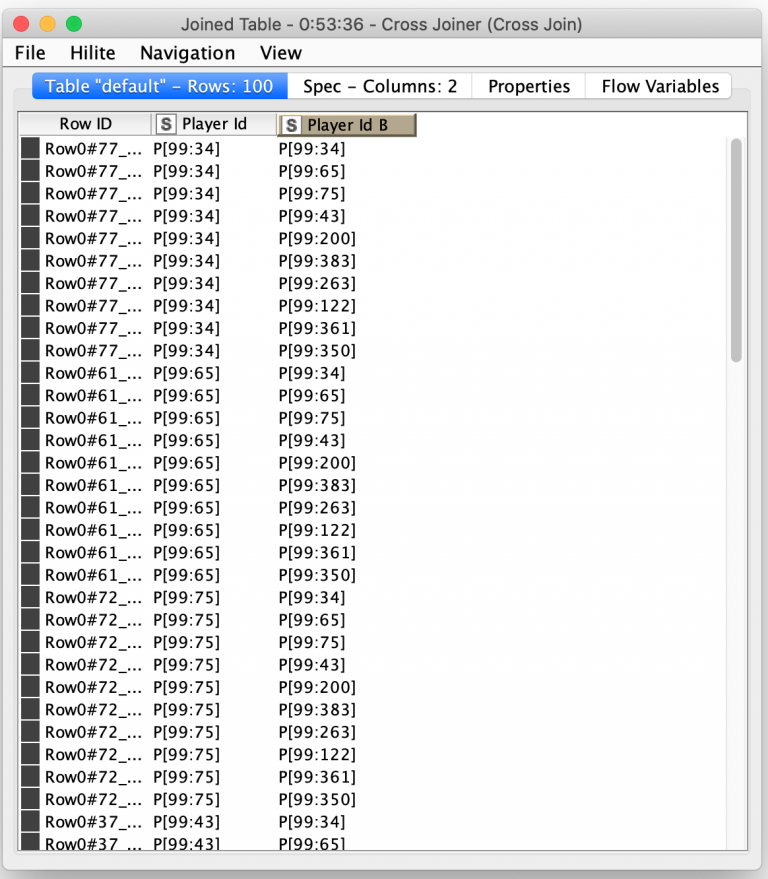
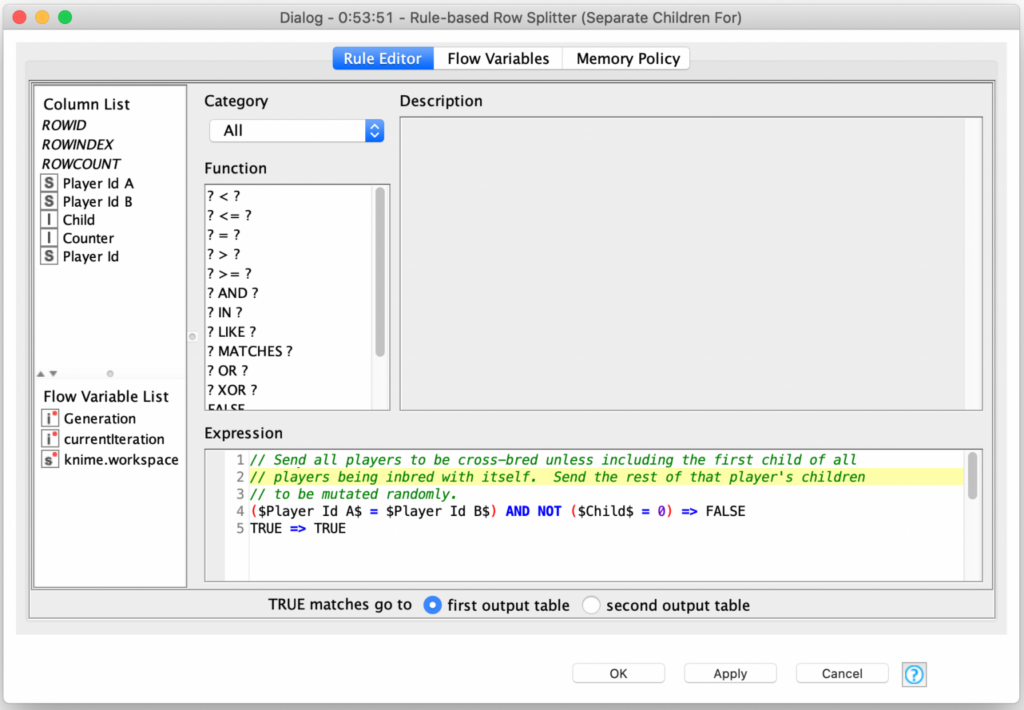
The combined are then cross joined again with a fixed size table of four rows that represent the offspring that the pair of parents will produce. The final output of these processing steps is 400 rows that each represent the parentage of the next generation of players. At this point, astute readers may have realized that the cross joiner will join a parent with itself and create four children for those records. If we are randomly selecting the traits from both parents, we will just end up with four identical copies of the parent. In biology, this is known as a clonal offspring. Rather than drop these records from consideration, I will instead use them during the final mutation phase of my genetic algorithm by keeping one child the same as the parent and randomly mutating the other three children.
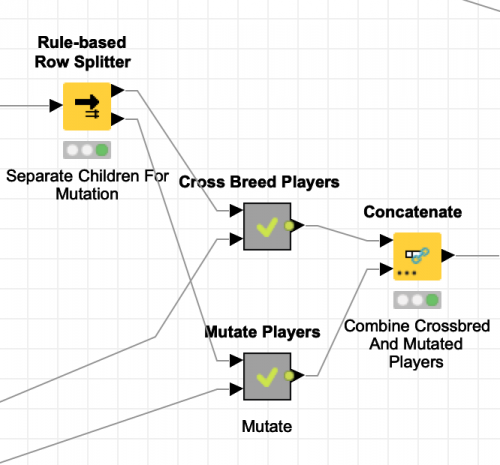
Which means that I now need to separate the offspring into two groups based on how they will be processed. The first group is the largest and consists of 370 children that will be cross bred (360 with different parents and 10 with the same parent). The remaining 30 children all have the same parent and are sent for mutation. This is easily accomplished with KNIME’s Rule-based Row Splitter node which allows for arbitrary expressions to control the selection of rows for each of its two output tables:
Cross breeding players for the next generation is surprisingly easy: we simply use the parent information to select each parent’s records and then arbitrarily pick rows from one parent or the other.
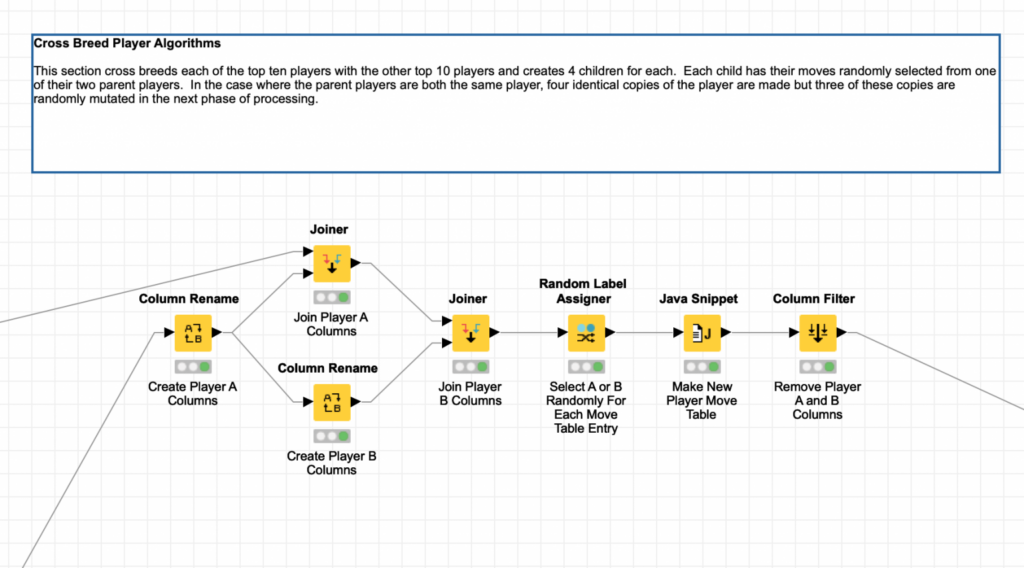
Again, KNIME has a node that makes this simple: the Random Label Assigner node. This node can be configured to randomly label each row of its input with a label based on a specified probability for the label. In this case, I simply want a 50/50 probability to pick the parent that a given row comes from:
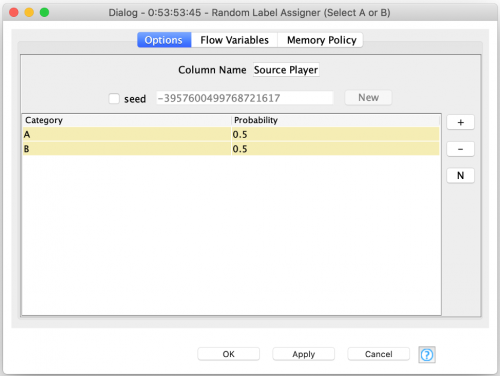
Once the source parent of each row is assigned, I use a Java Snippet to select that parent’s behavior for the child’s behavior table:
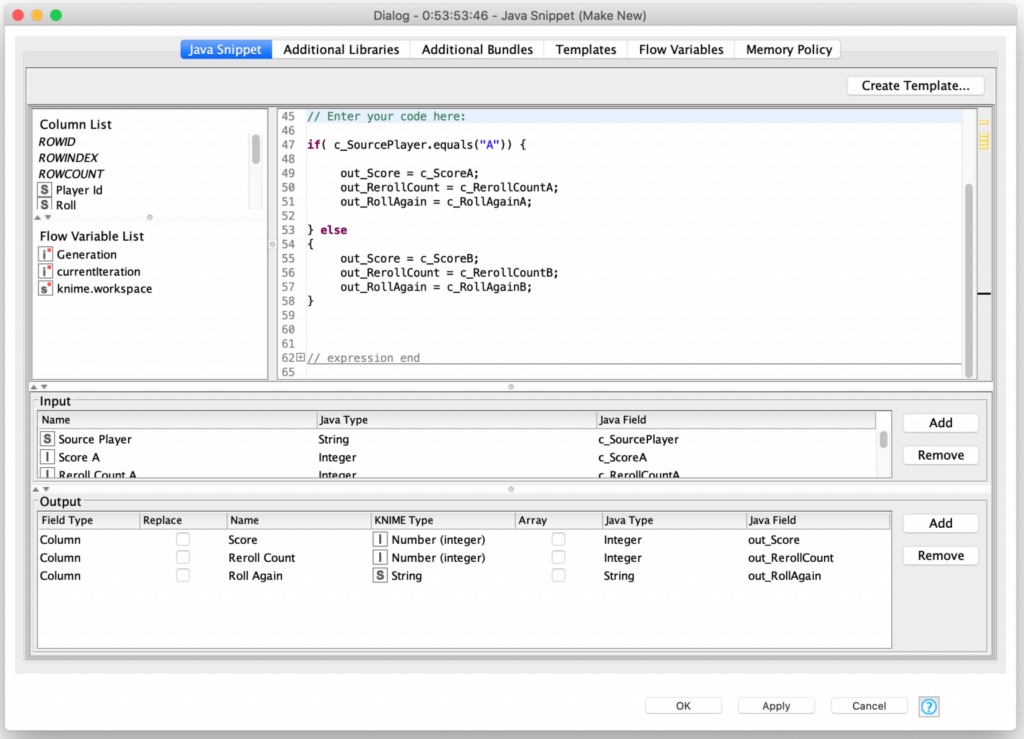
Mutation of the smaller group of remaining players (that have a clonal parent) is a bit more complex but it is very similar to the logic I used for creating the first generation of players randomly.
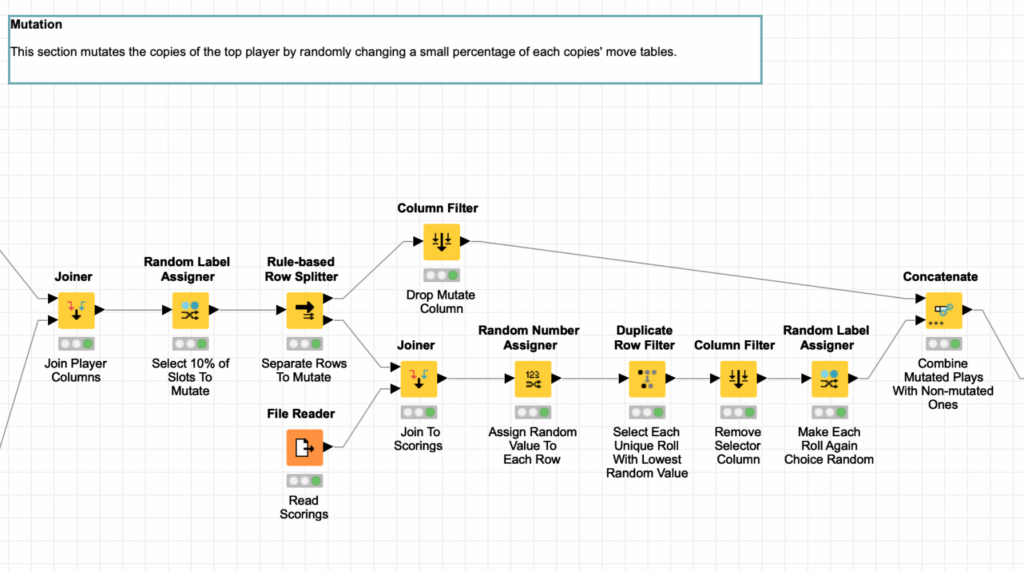
This process also uses the Random Label Assigner but instead leverages it to randomly select 10% of each child’s behavior table rows to be mutated.
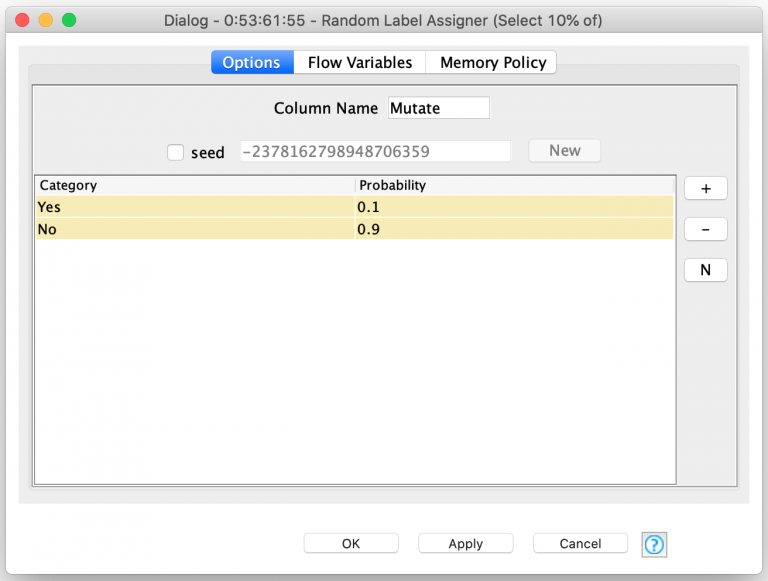
The rows that are labeled as requiring mutation are then replaced by a random corresponding row selected from the initial player template. Next, the roll again choice of the player is also randomly assigned for the new row. This technique is what I used to create the first generation of random players and I covered it in part two of this blog series.
Once all of the players have either been cross bred or mutated, they are recombined into a single table and this table is saved to disk as a single generation. This was done so that it is possible retrieve any given player and their ancestry which will also allow me to analyze the change of player behaviors over time. The new generation of players is then sent back through the algorithm to evolve the next generation using the Recursive Loop node as discussed in the previous blog posting.
While they are very powerful, there is a subtle danger lurking in the use of Recursive Nodes and my first version of my evolution workflow exposed this danger. When I first tried to evolve players, the performance of my workflow degraded with each new generation. It usually ground to a halt before the 10th generation and failed with an out of memory error. I was perplexed since the table size for each generation was the same. Suspecting some kind of memory leak in either the recursive or parallel loop nodes, I reached out to KNIME’s excellent engineering staff for help.
Fortunately, they quickly discovered the issue: in order to prevent collisions of the internal row identifiers for each generation, the Recursive Loop nodes append generational information to each row identifier. Because I leverage multiple nested sets of recursive loop nodes, my row identifiers were quickly growing extremely long and eventually exhausting the available heap space of the JVM that KNIME was running in. I had not noticed this because KNIME’s record viewer usually only shows the first few characters of the identifier column:
The fix was simple: I just added a RowID node at the end of my outermost loop to reset the row identifiers for the next generation. I appreciate the quick response from KNIME and their willingness to dig around in my workflow for the issue. Furthermore, they are considering product enhancements to both help detect long internal row identifiers and allow the loop nodes to be configurable to reset the identifiers after each iteration.
With my newfound ability to run as many generations as I wanted, I configured my workflow to try 100 of them and left it running overnight. The next morning, I was both excited and disappointed by the results:
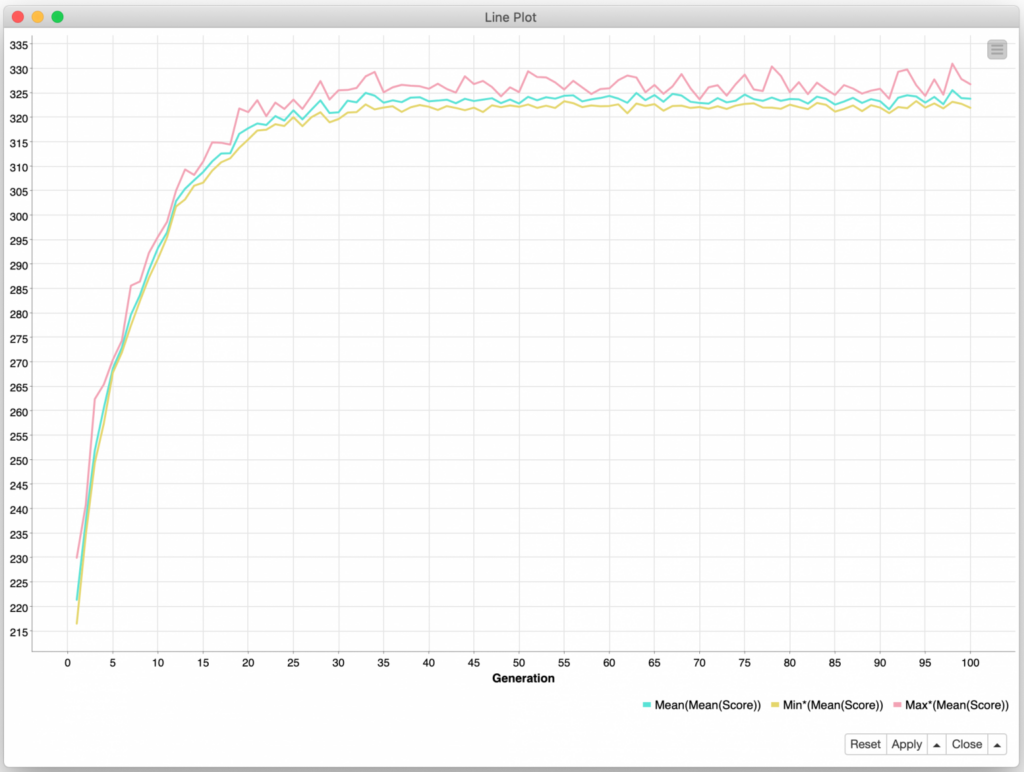
I was excited because the initial generations show a rapid and consistent improvement in average player score. But I was disappointed that the improvements leveled off by the 35th generation and showed no appreciable gains after that. While my overall approach has been validated, I believe the fitness function, selection criteria, and crossover/mutation operations all need further refinement to improve the ability of the overall algorithm to select and reinforce the optimal behaviors for the thousands of decisions that make up a single player’s “genetics”.
These potential improvements are outside of the scope of this blog series, but I find them intriguing enough to declare that I’m going to continue my explorations of the game of Farkle. This is the fundamental appeal of the game that has kept my wife and I so religiously playing it over the years: under a seemingly simple set of rules governed by basic combinatorics, there is a hidden complexity lurking.
And as for the player that all this evolutionary work produced? To put it kindly: sometimes evolution gives you a platypus. My wife, son, and I played several games where I used the move table of the highest average scoring player to control my decisions. I was soundly defeated in every game. While my prodigal player would make generally good moves on a turn by turn basis, its Achilles heel was the simplification I made early on in my modeling. I did not model the risk/reward calculation that a person would make as they accumulate a large score during an individual turn. My “single roll” rule-based player would continue rolling in situations a human most likely would not: e.g. despite having a large accumulation of points for the turn the player would continue to roll which would often lead to a Farkle and loss of all those potential points for that turn.
And so, I do plan to revisit Farkle in the future. I am going to build out a REST based game engine that will allow me to conduct head to head matches between both AI and human players. Perhaps there will be a Farkle tournament coming to a KNIME summit near you…

